What Are the New Findings?
The first reported confirmed SARS-CoV-2 infection in the U.S. was in Washington state on 21 Jan. 2020 in an individual who had traveled to Wuhan, China. To determine if SARS-CoV-2 infections were present in the U.S. prior to that date, samples from a DOD respiratory surveillance program dating back to 1 Dec. 2019, were evaluated for the presence of SARS-CoV-2 RNA. Viral RNA was not detected in any sample collected before 21 January 2020; however, positive samples were identified for several U.S. regions where the date of collection preceded the first reported infection for that region.
What Is the Impact on Readiness and Force Health Protection?
The implementation of new and emerging pathogen detection assays into already established surveillance programs could detect community spread earlier, thereby reducing the spread of pathogen among vulnerable populations more effectively.
Abstract
The first U.S. case of non-travel-related severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection was detected in late Feb. 2020 in California, but the prevailing delay in diagnostic testing and initial stringent testing criteria made it difficult to identify those who could have acquired the virus through community spread. The emergence of the virus in the western Pacific region in late 2019 and the global distribution of Department of Defense (DOD) personnel present the risk that DOD members may have been exposed and contracted the virus earlier then U.S. detections. Here, a retrospective study from residual samples collected from a global DOD Respiratory Surveillance Program was conducted to establish a tentative timeline of when this virus began circulating in the DOD population. Quantitative real-time reverse-transcription polymerase chain reaction testing for SARS-CoV-2 was performed and the specimen collection dates of positive results were compared to the dates of the first infections previously identified in respective states and counties. Twenty-four positive samples were identified out of approximately 7,000 tested. Although this retrospective testing found early cases in 8 locations, there were no results indicative of circulation before late February.
Background
Although the first 2 decades of the 21st century were marked by the emergence of novel respiratory pathogens with pandemic potential (2003 SARS-CoV, 2009 Influenza A H1N1, and 2012 MERS-CoV), the recognition and spread of a highly infectious and unusually lethal strain of the human beta coronaviruses, i.e., severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), in late 2019 caught the world's health systems by surprise. The emergent nature of the coronavirus disease 2019 (COVID-19) pandemic resulted in most of the scientific literature being released through the medRxiv preprint server prior to peer-review. As such, the authors recommend critical consideration of the referenced literature. Even the most technologically advanced countries with the greatest wealth, such as the U.S., did not have sufficient pandemic preparedness plans in place to adapt to this outbreak.1 In fact, the lack of centralized planning was evident in the wide variation between states on recommendations for testing in the early outbreak period between March (when the virus first arrived in most states) and July 2020.2 Even the Department of Defense (DOD), with its numerous world-class infectious disease research institutes and clinical surveillance programs, was caught off-guard. However, the DOD quickly adapted and put into place a surveillance approach leveraging its established Upper Respiratory Surveillance program housed at the U.S. Air Force School of Aerospace Medicine (USAFSAM). This response resulted in a massive capability increase and the DOD has remained ready to execute its global mission.
The gaps in early testing led to widespread underestimation of infected individuals, with some early sampling results suggesting nearly 5% of undiagnosed people in the U.S. were seropositive for prior exposure to the virus.3 According to Kalish et al., this seropositivity rate translates to 5 undiagnosed infections per diagnosis between May and July 2020. An additional impact of the variable testing recommendations and the high infectivity of the virus was the emergence of local hotspots such as New Orleans during Mardi Gras,4 where only a few individuals imported the virus but many exported it elsewhere.
In order to better prepare for the next major outbreak, a more detailed and thorough understanding of the early emergence of this pandemic in the U.S. and DOD-associated populations is required. To that end, a retrospective analysis was performed of clinical remnants from patients who reported to military medical treatment facilities during the winter of 2019–2020 complaining of upper respiratory infections. This paper reports the findings from that study and implications for increasing the DOD's readiness for the next pandemic.
Methods
The Air Force Research Laboratory Institutional Review Board (Study number FWR20190037N) determined this study to be non-human subject research as part of a public health response activity and the study was conducted between March and June 2020.
Sample selection and testing
De-identified nasopharyngeal swab (NPS) samples were collected from a respiratory surveillance program that tracks influenza globally to aid in the yearly influenza vaccine development. Samples were stabilized in viral transport media prior to testing, covered the date range from 1 Dec. 2019 to 3 June 2020, and originated from 86 military medical treatment facilities (MTFs) around the globe. USAFSAM (a DOD Reference Laboratory) previously tested these samples for upper respiratory infections using the Luminex Respiratory panel (Austin, TX) before being stored at -80º C. The Reference Laboratory provided the following metadata: date of collection, MTF, and co-infections.
Ribonucleic acid (RNA) was extracted from the NPS samples with Promega Maxwell 16 instruments using the Maxwell Total Viral RNA kit according to the manufacturer's instructions. Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) testing was performed using the SuperScript III RT-PCR master mix (ThermoFisher, cat. #204454) with the research use only 2019-nCoV primer-probe kit (IDT DNA, cat. #10006605). Thermocycling conditions on the ABI 7500 FAST analyzer consisted of a 20-minute reverse transcription step at 50° C, a 10-minute hot-start activation step at 95° C, and 45 cycles of 95° C for 3 seconds followed by 55° C for 30 seconds.
Data analysis and statistics
In accordance with the approved Centers for Disease Control and Prevention (CDC) assay methodology, samples were positive for SARS-CoV-2 if the RNase P control passed (cycle threshold [Ct] < 40) and both primer sets N1 and N2 produced Ct values below 40. In the cases where RNase P did not amplify but both N1 and N2 were positive, those samples were called positive in accordance with the Food and Drug Administration-authorized instructions for use of the CDC 2019-nCoV RT-PCR Diagnostic Panel. Samples where only 1 of the 2 markers were detected were "inconclusive" and the test was repeated using the previously extracted RNA. Sample metadata and amplicon data, which had been stored separately, were combined by the Laboratory Director prior to analysis. MS Access 2013 (Microsoft Corporation, Redmond, WA) was used for data management and descriptive statistics such as number of positives, daily positive hit rate, and earliest detection.
Results
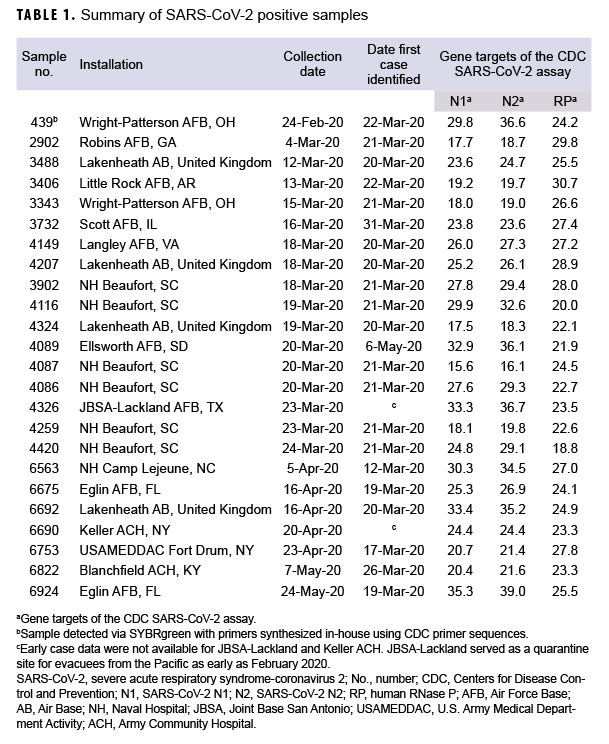
The observed positivity rate was 0.3% (24 positive samples from 7,021 total samples). Of these samples, 14 were collected from patients prior to the first COVID-19 case clinically reported at 8 different installations (Table 1). The first positive RT-PCR sample from Wright Patterson Air Force Base (AFB) in Ohio was collected a full month before the first laboratory-confirmed case at that installation (22 March 2020), and 2 days before the first laboratory-confirmed case in the DOD (26 February 2020 by U.S. Forces Korea).6 A second PCR-positive specimen of significant interest was collected at Ellsworth AFB 46 days before the first case of COVID-19 reported from the installation in South Dakota.
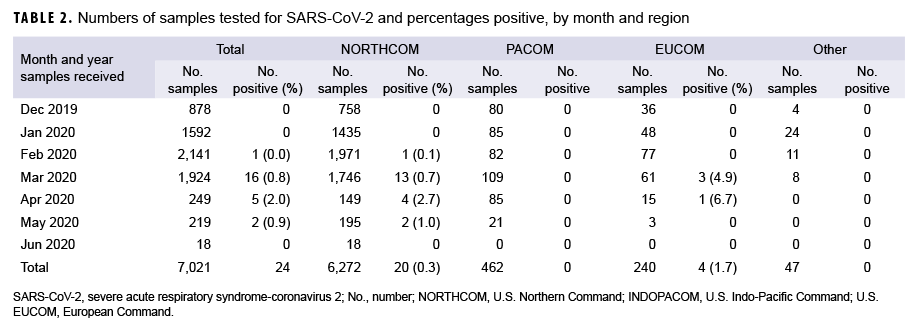
Samples were received from installations in 3 geographic combatant commands (Table 2): European Command (EUCOM), Pacific Command (PACOM), and Northern Command (NORTHCOM). The majority of test samples (89.3%) originated in NORTHCOM followed by PACOM (6.6%), and EUCOM (3.4%). An additional 47 samples originated in U.S. Coast Guard clinics (0.7%). Positive sample distribution included 20 from NORTHCOM (0.3% positive rate), 0 from PACOM, 4 from EUCOM (1.7% positive rate), and 0 from the Coast Guard.
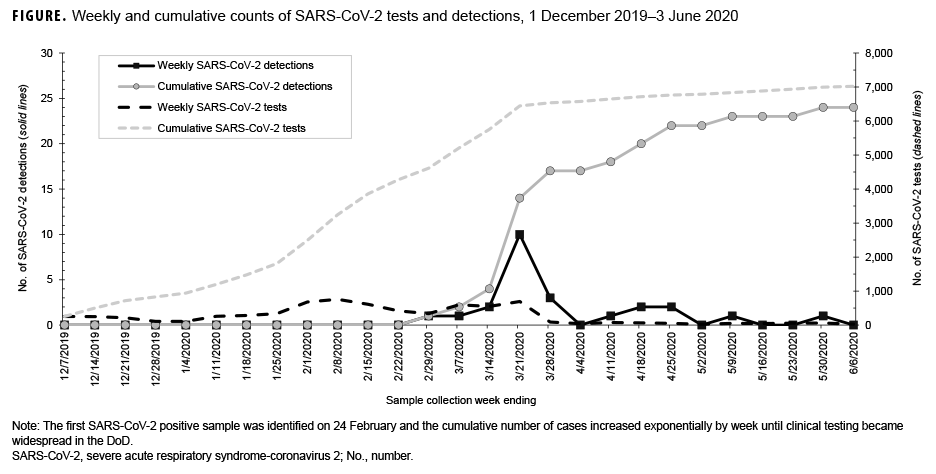
The number of SARS-CoV-2 positives in this respiratory surveillance sample set increased logarithmically with time, mirroring the positivity rate observed in the larger pandemic (Figure). Beginning in the week ending 29 Feb., an increasing number of positive tests was observed until the week ending 28 March. The decline at the end of March was due to the addition of SARSCoV-2 clinical testing in the Air Force clinical test menu in early March. Therefore, most of the SARS-CoV-2 positive samples in March and beyond were evaluated directly by the clinical lab and not by the surveillance lab. The effect of clinical testing was also seen in that the peak sampling period was from mid-February to early March, when nearly one-third of all samples were collected. Notably, the effect was most dramatically seen in testing numbers between the weeks ending 21 March and 28 March, when 691 samples were tested compared with 90, respectively.
In addition to observing an increase in testing, two peaks were detected in the positivity rates. For the week ending 29 Feb., one sample was positive out of 333 tests, a rate of 0.30%. The following two weeks remained below 1% positive rate (0.17% and 0.37%, respectively) before increasing to 1.45% with 10 detections out of 688 tests for the week ending 21 March, and peaking at 3.33% with three positive tests out of 90 samples during the final week of the study. The week ending 4 April had no positive tests, but then the next three weeks had positivity rates of 1.4%, 3.1%, and 3.8% before another week of no positive tests and finally a week with 2.0% positivity (1/49 tests). These observations are consistent with the observed increase in nationwide infection growth beginning during the week ending 14 March.7
Editorial Comment
These results suggest that while the COVID-19 virus was present in the U.S. military population earlier than previously reported, the change in observed infection timing was minimal. Excluding the outlier from Ellsworth AFB, the initial positive specimens were collected approximately 2 weeks before the first cases were reported (average=11.6 days, range 2–27 days). These results are similar to the results of other retrospective testing studies that focused on PCR-based detection of residual samples.8-10 Evaluation of residual specimens using antibody tests is still an on-going effort and may lead to different data as better assays become available.11
Collectively, the retrospective testing results worldwide demonstrate that the early testing response was inadequate. In the U.S., the lead for testing is the Centers for Disease Control and Prevention, guiding the development and deployment of diagnostic assays. In steady-state operations this is a fully acceptable arrangement; however, as has been seen during this outbreak, relying solely upon a single organization for direction has shortcomings. In contrast, the DOD's use of centralized control coupled with broad, decentralized execution provides the flexibility and adaptability that are necessary for early investigation of disease outbreaks.
When military planners return to the table to develop the next public health emergency preparedness plan, they should consider Joint doctrine and the wealth of expertise that the DOD has spread across its agency. The biological defense and public health enterprises centered around the Defense Threat Reduction Agency, the Defense Advanced Research Projects Agency, and the Armed Forces Health Surveillance Division utilize advanced molecular technology and deploy those capabilities across the services. Data from technologies such as next generation sequencing for detection could feed directly into the research labs so that diagnostics, vaccines, and therapeutics could be continually generated and transitioned to the industrial infrastructure at the first sign of outbreak. Technology is available and in wide use today that was not available in Dec. 2019 for detecting the earliest positives, but even with these advances in technology, as the results reported here demonstrate, outbreaks could be identified a matter of a few weeks earlier. For the next pandemic, the challenge is to have technology innovators engaged globally, and to adequately resource government, academic, and industry research labs to make a one month sequence-to-diagnostics turn-around a reality.
While more than 7,000 samples were tested in this study, the number of independent test sites is small and heavily favored by one Service. As expected a high proportion of samples originated from NORTHCOM facilities, where most of the DOD MTFs are located, while the overseas samples originated from 10 Air Force installations and 1 Army installation. In a demonstration of multi-agency collaboration, samples were received from 6 Coast Guard clinics, too. No samples were received from Southern Command and less than 10 samples from Central Command, so the impact of the epidemic in military members stationed in those regions is unknown. In order to provide adequate surveillance during normal operations and epidemics, the Military Health System, and the Defense Health Agency by extension, must increase the participation of sentinel sites in the Respiratory Surveillance Program. With 475 military hospitals and medical clinics across the globe, the DOD is the only health care system that is equipped to monitor worldwide infectious diseases before they enter the homeland. The surveillance network for the DOD is primed to immediately add validated assays for new and emerging infections in parallel with the clinical assays that are deployed, to actively track community spread once a virus has been detected by the CDC early in a pandemic.
Author affiliations: Department of Public Health and Preventive Medicine, U.S. Air Force School of Aerospace Medicine, Wright Patterson AFB, OH (all authors).
References
- Cirilo ER, Natti PL, Romeiro NML, et al. One study of COVID-19 spreading at the United States – Brazil – Columbia. medRxiv preprint, 29 Aug. 2020, https://doi.org/10.1101/2020.08.29.20184465
- Perniciaro SR and Weinberger DM. Variations in state-level SARS-CoV-2 testing recommendations in the United States, March-July 2020. medRxiv preprint 24 September 2020, https://doi.org/10.1101/2021.01.27.21250570
- Kalish H, Klumpp-Thomas C, Hunsberger S, et al. Mapping a pandemic: SARS-CoV-2 seropositivity in the United States. medRxiv preprint, 27 Jan. 2021, https://doi.org/10.1101/2021.01.27.21250570
- Zeller M, Gangavarapu K, Anderson C, et al. Emergence of an early SARS-CoV-2 epidemic in the United States. medRxiv preprint, 05 Feb. 2021, https://doi.org/10.1101/2021.02.05.21251235
- Centers for Disease Control and Prevention. Division of Viral Diseases. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel Instructions for Use. Revision 03. Catalog # 2019-nCoVEUA-01.
- U.S. Department of Defense. Coronavirus: DOD Response Timeline. Accessed 15 June 2020. https://www.defense.gov/Explore/Spotlight/Coronavirus/DOD-Response-Timeline/
- Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). COVID-19 Dashboard. Accessed 15 June 2020. https://coronavirus.jhu.edu/map.html
- Capalbo C, Bertamino E, Zerbetto A, et al. No Evidence of SARS-CoV-2 Circulation in Rome (Italy) during the Pre-Pandemic Period: Results of a Retrospective Surveillance. Int J Environ Res Public Health. 2020;17(22):8461.
- Yu X, Wei D, Chen Y, et al. Retrospective detection of SARS-CoV-2 in hospitalized patients with influenza-like illness. Emerg Microbes Infect. 2020;9(1):1470.
- Hogan CA, Garamani N, Sahoo MK, et al. Retrospective Screening for SARS-CoV-2 RNA in California, USA, Late 2019. Emerg Infect Dis. 2020;26(10):2487.
- Basavaraju SV, Patton ME, Grimm K, et al. Serologic testing of U.S. blood donations to identify SARS-CoV-2-reactive antibodies: Dec. 2019-Jan. 2020. Clinl Infect Dis. 2020;ciaa1785.